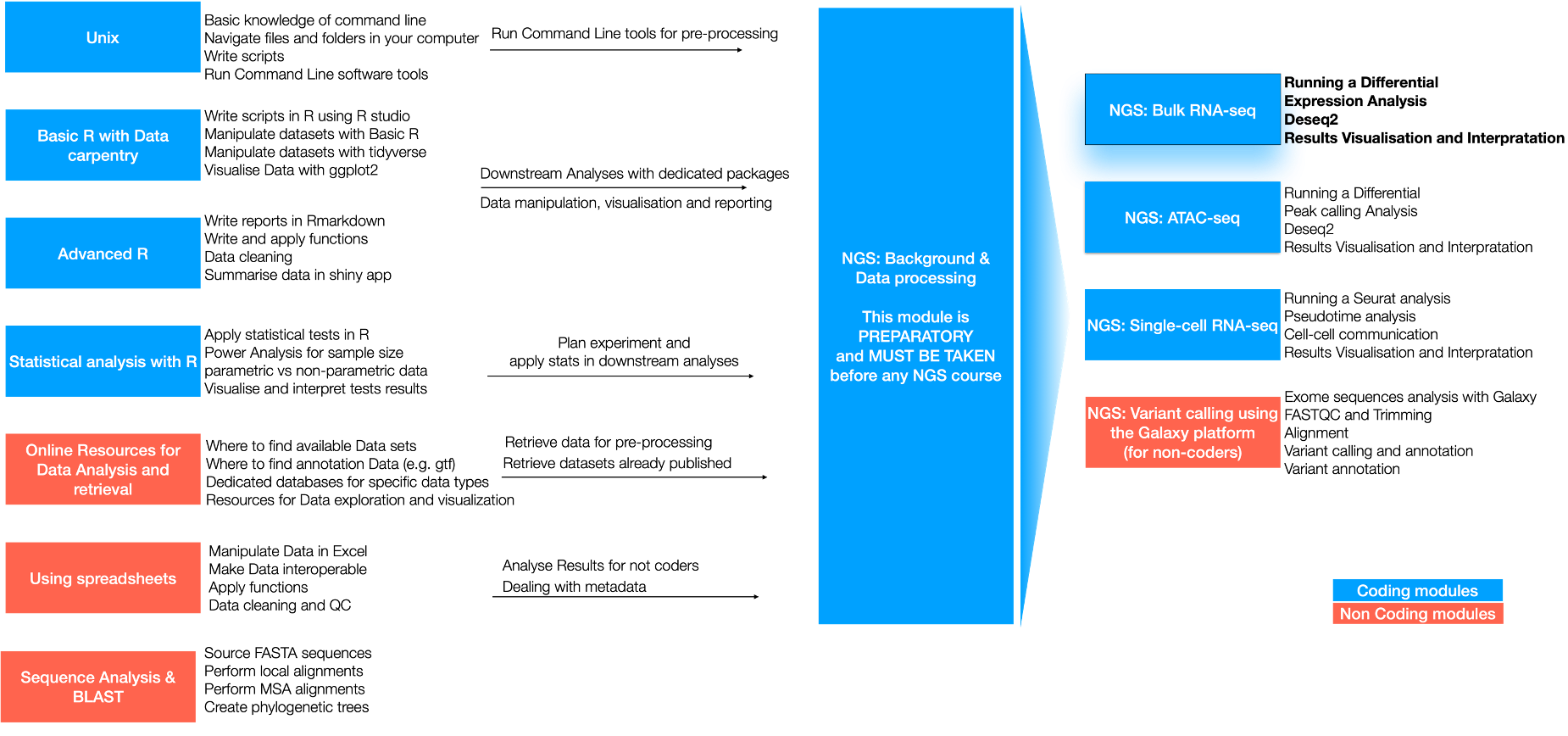
NGS: RNA-seq
Course Overview
-
Written by experts
-
Introductory
-
100% online
-
Video content
-
Multiple choice quiz
-
Approx. 2 hours to complete
-
Completion certificate
Write your awesome label here.
About this course
Course authors and designers
Dr. Alessandra Vigilante
Academic lead
Eva Hamrud
PhD Developmental Biology
Dr. Alex Thiery
Post-doc Bioinformatics
